Sunday, October 10, 2004 - 2:16 PM
6423
Gene Expression Patterns During Palatal Shelf Fusion: A Novel Approach to Identifying Genes Involved in Cleft Palate Formation
INTRODUCTION: Cleft palate is a common disorder that occurs when there is failure of the palatal shelves to fuse during normal development. The genes mediating normal palatal fusion are largely unknown.
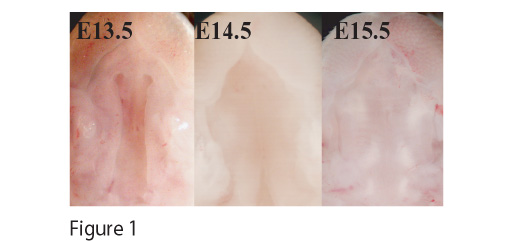
METHODS: Using microdissection techniques, we harvested mRNA and performed microarray (~40,000 element) analysis of mice palatal shelves at timepoints before, during, and after palatal fusion at E13.5, E14.5, and E15.5 (see Figure 1). We also performed histology and protein immunolocalization using standard techniques.
RESULTS: Replicates clustered together by timepoint indicating excellent consistency between palate harvests. Hox-7A, a gene implicated in both mice and humans as important to cleft lip and palate formation was upregulated during palatal fusion. Several retinoic acid responsive genes such as Stra6 were also upregulated during fusion. Furthermore, we found the homeobox gene Lim-1 to be upregulated during palatal fusion. To confirm our microarray findings of differential expression of LIM-1 during palate formation, we performed immunohistochemistry. During early secondary palate formation (E12.5), as the shelves are pointing inferiorly lateral to tongue, LIM-1 expression is maximal at the lateral wall and tip of the palate epithelia. Interstingly, minimal expression is noted in the medial palatal wall epithelia, facing the tongue. As the palatal fusion proceeds, LIM-1 expression is noted throughout the palatal epithelia. LIM-1 expression regress with epithelial to mesenchymal (EMT) transition at the midline epithelial seam as the approaching palates fuse on E14.5. Lim-1 expression is completely abolished within the palate mesenchyme by E16.5 after completion of fusion.
CONCLUSIONS: We have identified novel genes that may be involved in normal palatogenesis using large-scale microarray analysis. These exciting data may be a valuable resource for other researches attemping to elucidate the complex dysregulation of expression cascades involved in cleft palate formation. We hope to openly share these data with other cleft palate researches at the ASPS.
ACKNOWLEGEMENT: This work was generously supported by the PSEF and Smile Train Cleft Lip & Palate Research Initiative.
View Synopsis (.doc format, 138.0 kb)
See more of Cranio/Maxillofacial/Head & Neck
Back to 2004am Complete Scientific Program